Page Not Found
Page not found. Your pixels are in another canvas.
A list of all the posts and pages found on the site. For you robots out there is an XML version available for digesting as well.
Page not found. Your pixels are in another canvas.
This is a page not in th emain menu
Published:
This bit of code can be a great help to subset a Fasta file alignment based on certain conditions taken from a sample information file (e.g. subset only the samples collected in Spring, or those in Winter and greater than y cm, or everything except those in Autumn, etc.).
Published:
This post will show you how to generate a phylogeny plot in R, with a heatmap alongside it. This example uses binary data, but it can be adapted to be plotted with a continuous variable.
Published:
This R code shows you how to plot a map for a desired country or geographic region, and how to display a chosen climatic variable (e.g. mean annual temperature, annual precipitation) as an overlay. The WorldClim dataset has 19 variables to choose from.
Published:
This R script plots the map of a desired country, and colours it by elevation. It then adds GPS coordinate points to it (for example sampling sites) from an Excel file. Check out the final image at the bottom of this post 😃. The North arrow and Tetramesa wasp was added manually later in Inkscape.
Published:
I came across the problem of renaming sequences in a FASTA sequence alignment after downloading over 200 sequences from GenBank for four different genes. The sequence names were assigned by the GenBank accession number (e.g. MK1526) only. I wanted the sequence names to have the species name in it as well, for example MK1526_Canis_lupis. To avoid the tedious task of manually doing this, I wrote a few lines of R code that I hope will be of use to others with the same issue.
Published:
Perhaps a published paper lists their GenBank accession numbers as a range; for example KC664779 - KC665461. In R, one can use the following code to download all the sequences in this range, and save them as a FASTA file:
Published:
This snippet of Python or R code (whichever you prefer!) enables you to input a folder directory containing many FASTA files containing a single sequence in each, and combine them all into one FASTA file. I initially found it cumbersome to manually copy and paste the FASTA sequence from each trimmed chromatogram file into one big FASTA file. This automates the process, and could be useful to folks who are generating FASTA files for alignments 😃. This works correctly in Python 3.9. If you’re more of an R person, the R script is here too 😎.
An R Shiny application for processing binary data from fragment analysis methods such as ISSR and AFLP. 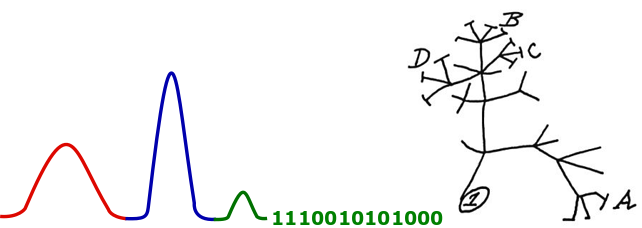
An R Shiny application to identify cochineal genetic sequences for 12S rRNA, 18S rRNA, or COI.
An R Shiny App that assesses the effects of methodological choice (e.g. tree prior, rate distribution, or clock rate) and sampling effects on the GMYC model for species delimitation.
Assesses sample size requirements for researchers performing critical thermal limits (CTL) studies
Published in Skeptical Inquirer, 2020
This article critically examines the claims in the book “Becoming Supernatural” by Joe Dispenza. 📁 PDF
Recommended citation: van Steenderen, C.J.M. 2020. Becoming Supernatural: How common people are being misled. Skeptical Inquirer Vol.44 No. 4. https://skepticalinquirer.org/authors/clarke-van-steenderen/
Published in Biological Control, 2021
Invasive Cactaceae cause considerable damage to ecosystem function and agricultural practices around the world. The most successful biological control agents used to combat this group of weeds belong to the genus Dactylopius (Hemiptera: Dactylopiidae), commonly known as ‘cochineal’. Effective control relies on selecting the correct species, or in some cases, the most effective intraspecific lineage, of cochineal for the target cactus species. Many of the Dactylopius species are so morphologically similar, and in the case of intraspecific lineages, identical, that numerous misidentifications have been made in the past. These errors have resulted in failed attempts at the biological control of some cactus species. This study aimed to generate a multi-locus genetic database to enable the accurate identification of dactylopiids. Genetic characterization was achieved through the nucleotide sequencing of three gene regions (12S rRNA, 18S rRNA, and COI) and two inter-simple sequence repeats (ISSR). Nucleotide sequences were very effective for species-level and D. tomentosus lineage-level identification, but could not distinguish between the two lineages within D. opuntiae commonly used for biological control of various Opuntia spp. Fragment analysis through the use of ISSRs successfully addressed this issue. This is the first time that a method has been developed that can distinguish between these two D. opuntiae lineages. Using the methods developed in this study, biological control practitioners can ensure that the most effective agent species and lineages are used for each cactus target weed, thus maximizing the level of control. 📁 PDF
Recommended citation: van Steenderen, C.J.M., Paterson, I.D., Edwards, S., and Day, M.D. 2021. Addressing the red flags in cochineal identification: The use of molecular techniques to identify cochineal insects that are used as biological control agents for invasive alien cacti. Biological Control 104426. doi: 10.1016/j.biocontrol.2020.104426. https://www.sciencedirect.com/science/article/pii/S1049964420306538
Published in Molecular Ecology Resources, 2022
Species delimitation tools are vital to taxonomy and the discovery of new species. These tools can make use of genetic data to estimate species boundaries, where one of the most widely-used methods is the Generalised Mixed Yule Coalescent (GMYC) model. Despite its popularity, a number of factors are known to influence the performance and resulting inferences of the GMYC. Moreover, the few studies that have assessed model performance to date have been predominantly based on simulated datasets, where model assumptions are not violated. Here, we present a user-friendly R Shiny application, “SPEDE-sampler” (SPEcies DElimitation sampler), that assesses the effect of computational and methodological choices, in combination with sampling effects, on the GMYC model. Output phylogenies are used to test the effect that 1) sample size, 2) BEAST and GMYC parameters (e.g. prior settings, single vs multiple threshold, clock model), and 3) singletons has on GMYC output. Optional predefined grouping information (e.g. morphospecies/ecotypes) can be uploaded in order to compare it to GMYC species and estimate percentage match scores. Additionally, predefined groups that contribute to inflated species richness estimates are identified by SPEDE-sampler, allowing for the further investigation of potential cryptic species or geographic sub-structuring in those groups. Merging by the GMYC is also recorded to identify where traditional taxonomy has overestimated species numbers. Four worked examples are provided to illustrate the functionality of the program’s workflow, and the variation that can arise when applying the GMYC model to empirical datasets. The R Shiny program is available for download on GitHub. 📁 PDF
Recommended citation: van Steenderen, C.J.M. and Sutton, G.F. 2022. SPEDE-sampler: an R Shiny application to assess how methodological choices and taxon-sampling can affect Generalised Mixed Yule Coalescent (GMYC) output and interpretation. Molecular Ecology Resources (22)2 doi: 10.1111/1755-0998.13591 https://onlinelibrary.wiley.com/doi/abs/10.1111/1755-0998.13591
Published in Biodiversity Data Journal, 2022
Processing and visualising trends in the binary data (presence or absence of electropherogram peaks), obtained from fragment analysis methods in molecular biology, can be a time-consuming and often cumbersome process. Scoring and analysing binary data (from methods, such as AFLPs, ISSRs and RFLPs) entail complex workflows that require a high level of computational and bioinformatic skills. The application presented here (BinMat) is a free, open-source and user-friendly R Shiny programme (https:// clarkevansteenderen.shinyapps.io/BINMAT/) that automates the analysis pipeline on one platform. It is also available as an R package on the Comprehensive R Archive Network (CRAN) (https://cran.r-project.org/web/packages/BinMat/index.html). BinMat consolidates replicate sample pairs of binary data into consensus reads, produces summary statistics and allows the user to visualise their data as ordination plots and clustering trees without having to use multiple programmes and input files or rely on previous programming experience. 📁 PDF
Recommended citation: van Steenderen, C.J.M. 2022. Biodiversity Data Journal (10) doi: 10.3897/BDJ.10.e77875 https://bdj.pensoft.net/article/77875/
Published:
The ability to use genetic barcoding tools to distinguish between the species and intraspecific ‘biotypes’ within the Dactylopius genus (Hemiptera: Dactylopiidae) is highly beneficial to the biological control of cactaceous weeds in South Africa. The present study used DNA sequencing and ISSR fragment analysis methods to create a database of genetic barcodes for Dactylopius species found in the country, as well as from the native range, with a particular focus on the biotypes found within Dactylopius opuntiae. This has important applications in the mass rearing of pure insect cultures and the inoculation of the most effective biotype on target Cactaceae.
Published:
Invasive Cactaceae cause considerable damage to ecosystem function and agricultural practices around the world but some cacti are also important and valued crop species. The most successful biological control agents used to combat cactus weeds belong to the genus Dactylopius (Hemiptera: Dactylopiidae), commonly known as ‘cochineal’, but the worst pests of cactus crops are also members of this genus. Cochineal lineages used for biocontrol of cactus weeds are host specific and only certain species and lineages will feed on cactus crops, so cactus biocontrol can be safely implemented without harm to cactus agriculture. Many of the Dactylopius species are so morphologically similar, and in the case of intraspecific lineages, identical, that numerous misidentifications have been made in the past. These errors may result in cactus farmers incorrectly assuming that the biocontrol agent is damaging their crop. This study aimed to generate a multi-locus genetic database to enable the accurate identification of dactylopiids. This was achieved through the nucleotide sequencing of three gene regions (12S rRNA, 18S rRNA, and COI) and two inter-simple sequence repeats (ISSR). Nucleotide sequences were very effective for species-level and D. tomentosus lineage-level identification, but could not distinguish between the two lineages within D. opuntiae commonly used for biological control of various Opuntia spp. Fragment analysis through the use of ISSRs successfully addressed this issue. This is the first time that a method has been developed that can distinguish between these two D. opuntiae lineages. Using the methods developed here, one can distinguish between what is a potential pest, and what is a beneficial biological control agent.
Published:
The Tetramesa genus (Hymenoptera: Eurytomidae) comprises at least 200 species that feed exclusively on grasses. The highly host-specific behaviour of these wasps, and the damage that they can cause to their host plants, makes them ideal biological control agent candidates for invasive grasses. Very little is known about the Afrotropical Hymenoptera in general, and to date, almost all of the sampling effort in collecting and describing Tetramesa species has taken place in the Northern Hemisphere. Only four African species have been described; none of which are from South Africa. The Centre for Biological Control (CBC) at Rhodes University has been investigating biological control options for several African grasses that have become invasive in Australia and the Americas, and have been collecting Tetramesa specimens across South Africa since 2017. The insect communities associated with more than 55 different native grasses have been surveyed over this period. The uniform morphology of adult and larval Tetramesa has, however, made it impossible to determine whether these wasps are a single polyphagous species, or multiple oligophagous and/or monophagous species. We are currently using genetic barcoding tools (mitochondrial COI and nuclear ITS2 regions) and species delimitation methods to solve this problem. Our preliminary results have identified at least four putative species (or rather ‘molecular operational taxonomic units’ (MOTUs)). These were collected from single host plants, confirming their host-specificity and potential as biological control agents. It is likely that we will uncover many more undescribed species in the region as our sampling effort escalates.
Published:
The stem-boring wasp genus Tetramesa (Hymenoptera: Eurytomidae) comprises 203 species that feed exclusively on grasses. The wasps are highly host-specific, typically feeding on a single or a few closely-related grasses, and can cause significant damage to their host grass (e.g. reducing seed production, increasing tiller mortality). These attributes often result in Tetramesa being serious grain pests, but it also makes them ideal biological control agent candidates for controlling invasive grasses. Very little is known about the Afrotropical Hymenoptera in general, and to date, almost all the sampling effort in collecting and describing Tetramesa species has taken place in the northern hemisphere. Only four African species have been described; none of which are from South Africa. The Centre for Biological Control (CBC) at Rhodes University has been investigating biological control options for several African grasses that have become invasive in Australia and the Americas, and have been collecting Tetramesa specimens across South Africa since 2017. The insect communities associated with more than 60 different native grasses have been surveyed over this period. The uniform morphology of adult and larval Tetramesa has, however, made it impossible to determine whether these wasps are a single polyphagous species, or multiple oligophagous and/or monophagous species. We are currently using genetic barcoding tools (mitochondrial COI and nuclear ITS2 regions) and species delimitation methods to solve this problem. Our preliminary results have identified at least six potentially undescribed Tetramesa species from South Africa. Each novel Tetramesa species was highly specific, with five of the six potential species feeding and completing their development on a single host grass species. This work will facilitate using biological control techniques to manage invasive alien grass species and highlights a previously unknown diversity of Tetramesa species associated with South African grasses. It is likely that we will uncover many more undescribed Tetramesa species in the region as our sampling effort escalates.
, Rhodes University, Department of Zoology and Entomology, 2021
TBA